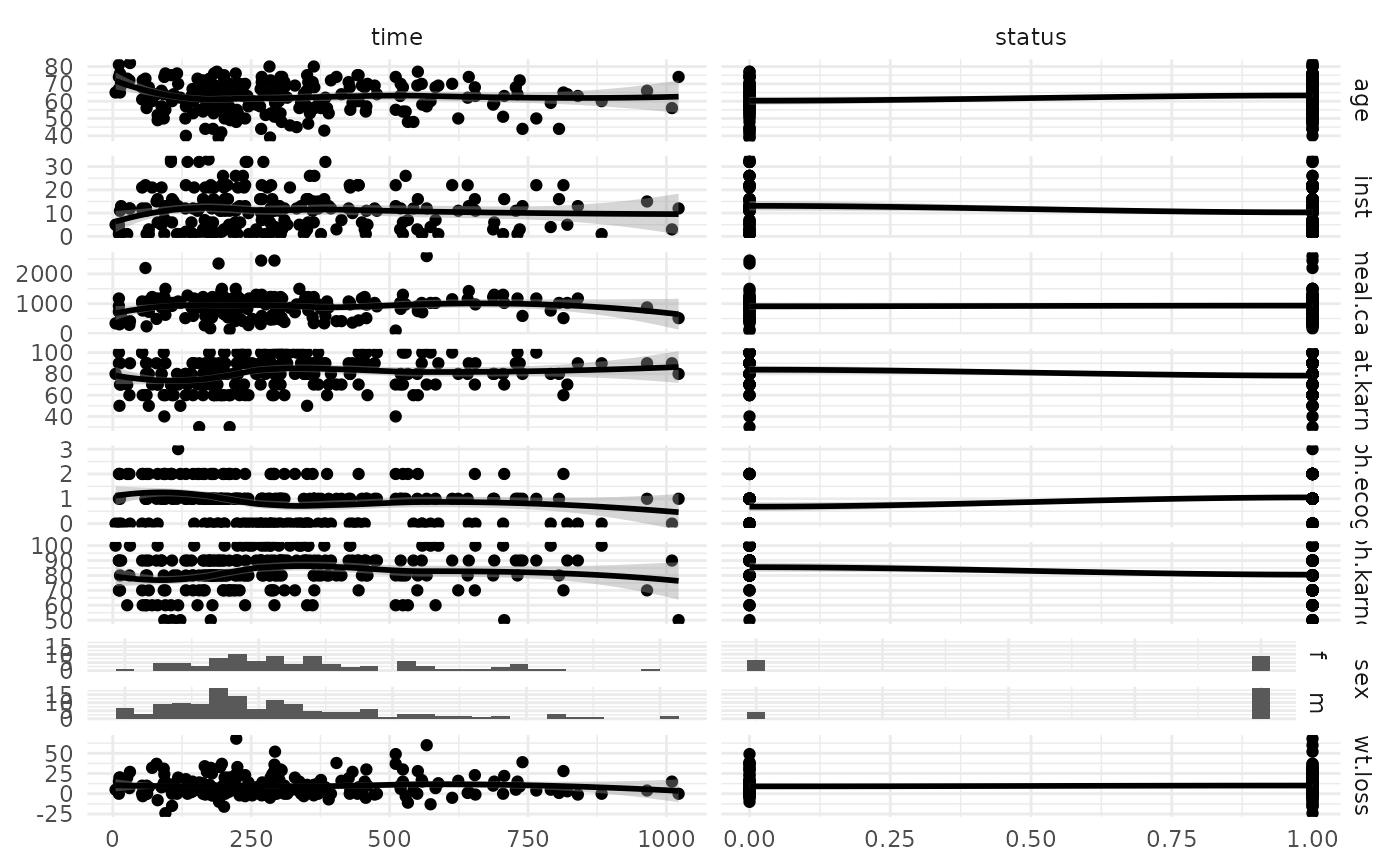
Generates plots for TaskSurv, depending on argument type:
"target": CallsGGally::ggsurv()on asurvival::survfit()object. This computes the Kaplan-Meier survival curve for the observations if this task."duo": Passes data and additional arguments down toGGally::ggduo().columnsXis target,columnsYis features."pairs": Passes data and additional arguments down toGGally::ggpairs(). Color is set to target column.
Usage
# S3 method for class 'TaskSurv'
autoplot(
object,
type = "target",
theme = theme_minimal(),
reverse = FALSE,
...
)Arguments
- object
(TaskSurv).
- type
(
character(1))
Type of the plot. See Description for available choices.- theme
(
ggplot2::theme())
Theggplot2::theme_minimal()is applied by default to all plots.- reverse
(
logical())
IfTRUEandtype = 'target', it plots the Kaplan-Meier curve of the censoring distribution. Default isFALSE.- ...
(
any)
Additional arguments.rhsis passed down to$formulaof TaskSurv for stratification for type"target". Other arguments are passed to the respective underlying plot functions.
Value
ggplot2::ggplot() object.
Examples
library(ggplot2)
task = tsk("lung")
task$head()
#> time status age meal.cal pat.karno ph.ecog ph.karno sex wt.loss
#> <int> <int> <int> <int> <int> <int> <int> <fctr> <int>
#> 1: 455 1 68 1225 90 0 90 m 15
#> 2: 210 1 57 1150 60 1 90 m 11
#> 3: 1022 0 74 513 80 1 50 m 0
#> 4: 310 1 68 384 60 2 70 f 10
#> 5: 361 1 71 538 80 2 60 f 1
#> 6: 218 1 53 825 80 1 70 m 16
autoplot(task) # KM
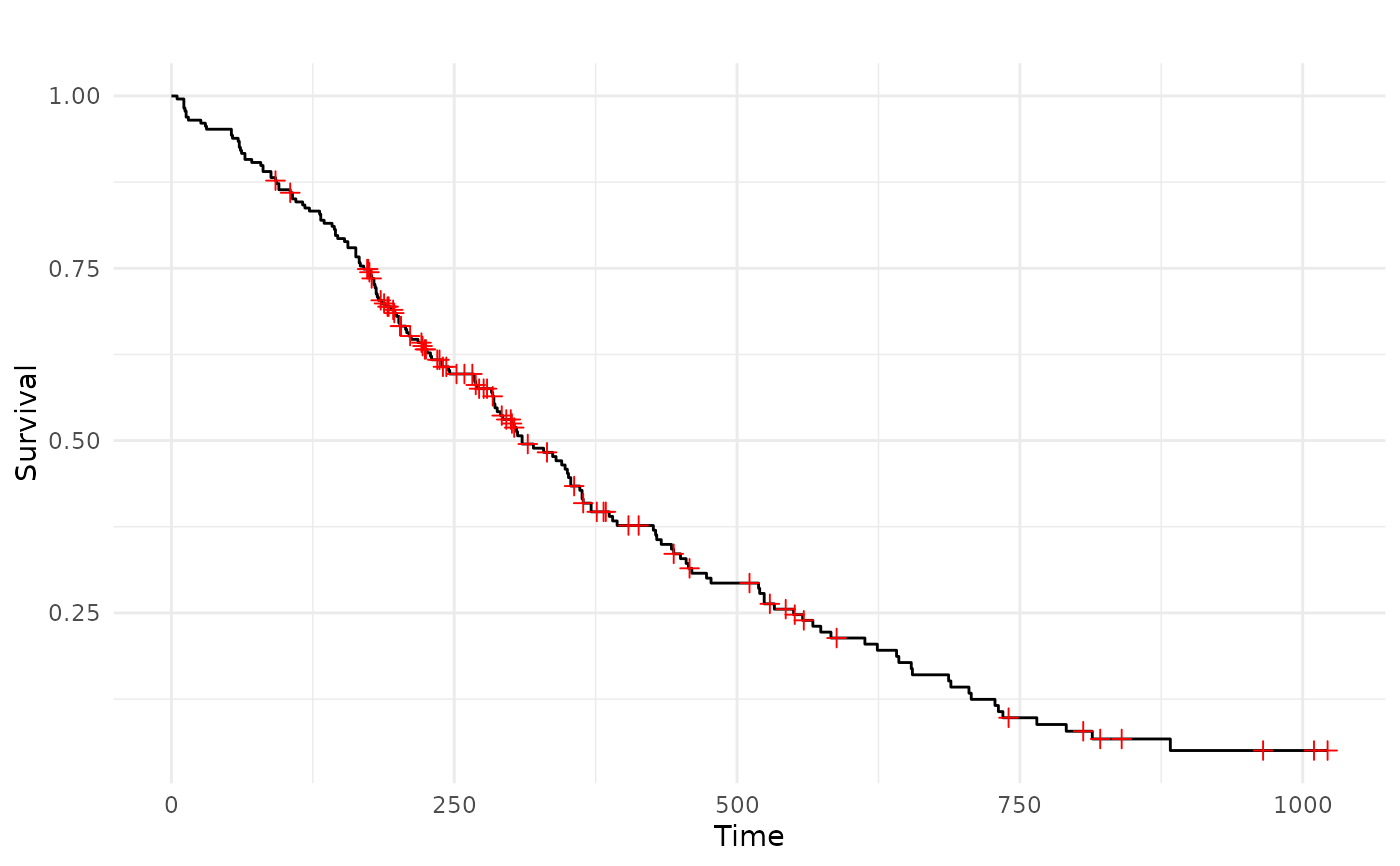 autoplot(task) # KM of the censoring distribution
autoplot(task) # KM of the censoring distribution
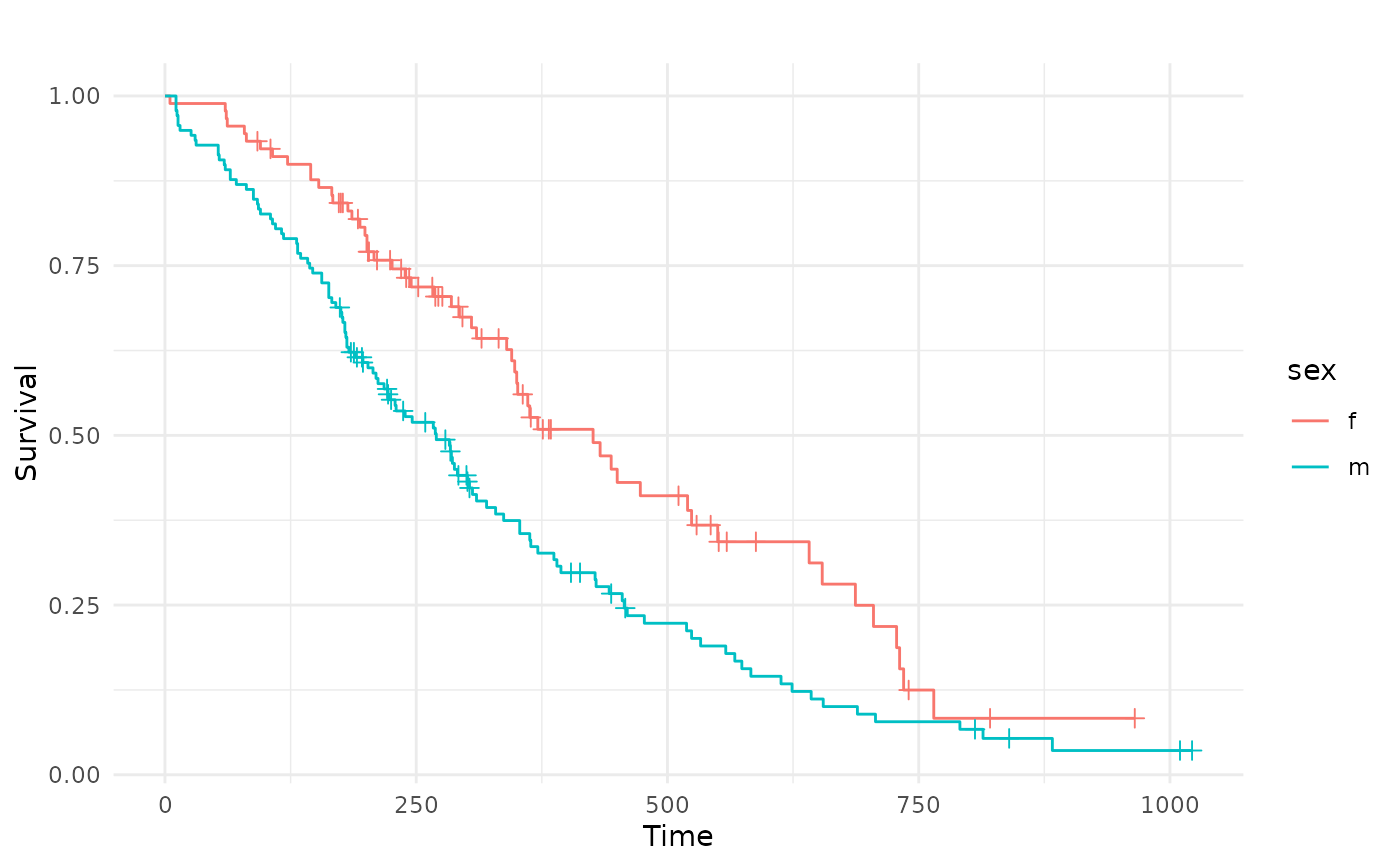
 autoplot(task, rhs = "sex")
autoplot(task, rhs = "sex")
 autoplot(task, type = "duo")
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
autoplot(task, type = "duo")
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01
#> Warning: pseudoinverse used at -0.005
#> Warning: neighborhood radius 1.005
#> Warning: reciprocal condition number 0
#> Warning: There are other near singularities as well. 1.01